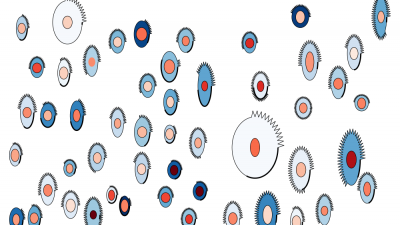
Modeling and visualization of receptor clustering on the cellular membrane
This novel mathematical model describes the stochastic process of the ligand-receptor clustering. To study the structure and the size of the ligand-receptor clusters, a stochastic particle simulation is employed. Besides the translation of the particles on the cellular membrane, the particle rotation is taken into account as binding sites are explicitly modelled. Glyph-based visualization techniques are used to validate and analyze the results of our in-silico model. Information on the individual clusters as well as particle-specific data can be selected by the user and is mapped to colors to highlight certain properties of the data. The visualization supports the process of model development by visual data analysis including the identification of cluster components as well as the illustration of particle trajectories.
| Release Date: | |
| Status: | |
| Availability: | |
| Data type: | |
| Techniques: | 2D, 3D, Spatial representation |
| Software: | |
| Technology: | C++ and OpenGL |
| Platform: | |
| Requirements: |
Project development
The project was completed with the support of The German Research Foundation (DFG), within the Cluster of Excellence in Simulation Technology (EXC 310/1) at the University of Stuttgart.
Last updated on 5th January, 2017
- Log in to post comments